pairs2groups¶
find homogeneous groups of items based on pairwise information
A motivating example¶
Suppose you have performed several experiments from four treatments (treatments 1,2,3, and 4). From each treatment, you have collected many independent samples. The question is into what groups of ‘statistically not different’ the results may be divided.
This package, pairs2groups, finds the groups into which the treatments may be divided such that each member of a group is not significantly different than other members in the group.
A picture may make this more clear. Samples from each treatment are plotted below in a boxplot. The letters below each box describe the groups each treatment is a member of. For example, all treatments in group ‘a’ are not statistically significantly different than each other.
In this example, treatment 1 is in groups ‘a’ and ‘b’. Therefore, it is statistically significantly different from treatment 4, which is not in either of these groups. Treatment 1 is not statistically significantly different from treatments 2 or 3, which which it shares membership in groups ‘b’ and ‘a’, respectively.
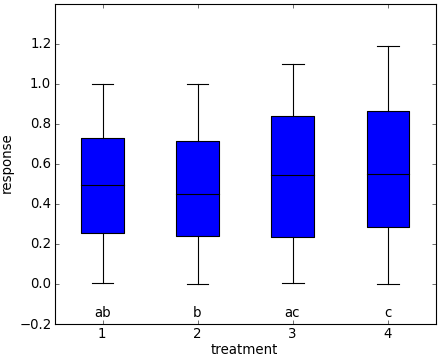
Problem definition¶
The problem is how to find the (minimal but complete) set of homogeneous groups of a collection of items. A homogeneous group is defined to have no member that is ‘different’ (defined below) than any other member.
Consider the problem of n items and pairwise knowledge of whether each item is either ‘different’ or ‘not different’ from every other item. This property of ‘different’ is commutative (A is different than B means B is different than A), but not transtive (A is different than B is different than C does not specify the relation between A and C).
How to construct groups such that every member population of a group is not different than the other populations in the group?
Development information¶
The source code and issue tracker for this library are at http://github.com/astraw/pairs2groups
Top-level functions¶
- pairs2groups.label_homogeneous_groups(populations, significance_level=0.050000000000000003, two_tailed=True, force_letter=True)¶
perform statistical comparisons and call find_homogeneous_groups()
The statistical test used is the Mann Whitney U.
Parameters: populations : A sequence of of sequences
The list of populations to analyze.
significance_level : float, optional
The significance level required to determine two groups different.
two_tailed : bool, optional
Whether the comparison is two-tailed.
force_letter : bool, optional
If true (the default), each population gets assigned a letter
Returns: group_info : dictionary
These keys are returned: ‘groups’ specifies the population indices for each group, ‘group_strings’ assigns letters to each group and returns a list of strings for each item, ‘p_values’ is a matrix of p values, ‘medians’ is the median of each population.
Examples
This example generates four populations. Three from the same distribution, and the last from a different distribution. Then, label_homogeneous_groups() is used to find which of these populations belong to statistically non significantly different groups.
>>> import numpy as np >>> pop1 = np.random.normal(size=(100,)) >>> pop2 = np.random.normal(size=(100,)) >>> pop3 = np.random.normal(size=(100,)) >>> pop4 = np.random.normal(size=(100,)) + 2 >>> populations = [pop1, pop2, pop3, pop4] >>> group_info = label_homogeneous_groups(populations) >>> group_info # doctest: +SKIP {'p_values': array([[ NaN, 0.578, 0.705 , 0. ], [ 0.578, NaN, 0.855, 0. ], [ 0.705 , 0.855, NaN, 0. ], [ 0. , 0. , 0. , NaN]]), 'medians': [0.071, -0.010, -0.0156, 2.054], 'group_strings': ['a', 'a', 'a', ''], 'groups': [(0, 1, 2)]}
- pairs2groups.find_homogeneous_groups(different_pairs, N_items)¶
Find all homogeneous groups of not-different pairs.
The algorithm used is as follows, where S is the set of all n items.
- Set k equal n, and T equal S.
- Set m equal n choose k. Take all (m in number) k element subsets of T. Denote the i*th subset of *T as U_i.
- For i in (0, ..., m-1):
- 3a. If no pair within U_i is different, then U_i is a
- group. Remember it.
3b. Else, set k equal k-1, and T equal U_i. Goto 2.
Parameters: different_pairs : list of 2-tuples
A list of pairs specifying different between two items
N_popupations : int
The number of items in the population
pairs2groups is a software package for Python.
Running the unit tests¶
After installation, do:
nosetests pairs2groups.util pairs2groups -v -v --with-doctest
Utility functions¶
- class pairs2groups.util.UnorderedPair¶
a 2-tuple that hashes the same regardless of order
Methods
- pairs2groups.util.get_all_pairs(S)¶
get a list of all pairs of values of S
>>> [tuple(p) for p in get_all_pairs( [0,1,2,3] )] [(0, 1), (0, 2), (0, 3), (1, 2), (1, 3), (2, 3)]
- pairs2groups.util.get_k_element_subsets(k, T)¶
get all subsets of T of length k
Returns len(T) choose k subsets.
- pairs2groups.util.is_list_of_sets_equal(A, B)¶
test whether list of sets A is equal to list of sets B
>>> A = [frozenset([1])]
>>> B = [frozenset([1])] >>> is_list_of_sets_equal(A, B) True
>>> B = [frozenset([2])] >>> is_list_of_sets_equal(A, B) False
>>> B = [frozenset([1]),frozenset([2])] >>> is_list_of_sets_equal(A, B) False
>>> B = [] >>> is_list_of_sets_equal(A, B) False
- pairs2groups.util.remove_overlapping_subsets(S)¶
- given a list of sets S, find the minimal list T of unique sets such that every set in S is a subset of a set in T.
- pairs2groups.util.take_not(T, j)¶
remove jth element of T
>>> T = [0, 1, 2, 3] >>> take_not( T, 1) [0, 2, 3]